Volume 4, Issue 4 (Autumn 2015)
J Occup Health Epidemiol 2015, 4(4): 229-240 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Safaei A, Azad M, Abdi F. A suitable data model for HIV infection and epidemic detection. J Occup Health Epidemiol 2015; 4 (4) :229-240
URL: http://johe.rums.ac.ir/article-1-167-en.html
URL: http://johe.rums.ac.ir/article-1-167-en.html
Related article in
Google Scholar
Google Scholar
Similar articles
1- Dept. of Medical Informatics, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran , aa.safaei@modares.ac.ir
2- Dept. of Computer Engineering, Qeshm International Branch, Islamic Azad University, Qeshm, Iran.
3- Nima Institute, Mahmoodabad, Mazandaran, Iran.
2- Dept. of Computer Engineering, Qeshm International Branch, Islamic Azad University, Qeshm, Iran.
3- Nima Institute, Mahmoodabad, Mazandaran, Iran.
Article history


Received: 2015/04/26
Accepted: 2015/10/9
ePublished: 2015/12/29
Accepted: 2015/10/9
ePublished: 2015/12/29
Subject:
Epidemiology
Full-Text [PDF 555 kb]
(1885 Downloads)
| Abstract (HTML) (7824 Views)
Table 1: Relational database
.jpg)
Figure 1: An overview of the Entity Attribute Value (EAV) (12)
.jpg)
Figure 2: The relationship between two nodes and human attributes on the relational edge
.jpg)
.jpg)
Figure 4: Comparison of graph database and relational database in terms of storage space occupation
Table 2: Comparison of the data retrieval time for different queries in the graph data model and the relational data model
Another issue is finding the source of infection. To achieve this goal, it is necessary to find the infection path. In the proposed graph data model, this would be gained simply by tracking the edges of infection, while in the relational data model, many regression procedures are needed which make it difficult to use. The time consumed to find the infection path to the level of four was 45 milliseconds for the graph model, and 920 milliseconds for the relational model. By increasing the level of infection path, the performance of the graph model increased compared to the relational model.
One of the important queries in this field is finding people who are at risk of the disease. To answer this question, it is necessary to find individuals located in the neighborhood of the infected individuals or at a certain distance from them. In the graph model, this only requires tracking to the desired depth of the graph. However, in the relational model, a connection to a large number of tables is needed and this takes a long time. For example, in the graph data model, it takes 8540 milliseconds to achieve a depth of four, while it takes 2354921 milliseconds in the relational data model (about 276 times longer than the required time in the graph data model). Moreover, the relational model cannot respond to tracking of more than level four.
Another feature of this graph data model is the capability to display all the nodes, edges, and attributes at the same time (row 4 of table 2), but this feature is not available in relational databases.
Discussion
As was mentioned, diseases have been of critical concern to societies and their manpower. Thus, it is essential to accumulate information about diseases and their preventive methods, especially viral diseases. To this purpose, a structure is required to store data in a specific format by which correct analysis is possible. In this study, the graph structure (data model) was used to store data, in which individuals’ features are recorded in the nodes as attributes and edges represent social relationships between them. In the case of transmission of infection from one person to another, the infection attribute becomes "true" for them. After identifying the disease, other entities with their own features and attributes will be added; these entities include infection path, physician, and medication. This model has high performance in answering the queries related to individuals’ relationships such as the disease infection path and people at risk of the infection. Nevertheless, the relational model has a better function only in queries related to data retrieval.
This system can be improved by the combination of the graph model and other models so that it would be able to effectively answer both types of queries (retrieval of individuals’ features and tracking of their relationships). For example, the combination of graph and relational models is an appropriate choice to store network epidemic data. Moreover, the focus of this study was static social networks. Hence, it is important to offer a model to store dynamic social networks such as individuals’ communication patterns, time, and location.
Conclusion
The proposed system can quickly provide the time and onset of disease transmission on different levels of individuals’ relationships. It makes possible to determine the one beginning point of disease transmission in a specific region of society, and the way by which transmission has occurred. These reports and related analyses can effectively help prevent disease incidence and viral transmission route in epidemic diseases. Using the graph data model allows us to make changes in information at running time, even if the necessary data are not considered while designing a process which is not feasible in other systems.
The management of data for epidemic detection of HIV infection requires an appropriate data model that can provide the required functionalities and features with an acceptable quality. Graph data models are suitable NoSQL models for some of these features (e.g., epidemic detection via traversing of the graph). The proposed graph-based data model provides the main functionalities and features while outperforming performance and utilization metrics.
Acknowledgments
We would like to thank our colleagues at Tarbiat Modares University and Islamic Azad University (Qeshm Island Unit).
Conflict of Interest: None declared.
Full-Text: (259 Views)
Introduction
One of the issues that communities and organizations are facing is the incidence of new diseases, methods, and changes which significantly affect the health of the society. These changes and the way they are dealt with strongly impact the success and failure of the society. Since the main factor in health care is prevention and quick action after the outbreak of disease, automating these actions would play a significant role in improving the performance of health care providers. Technological changes and disease outbreak are two important issues for governments, the lack of adaptation to which would be harmful to the community, and the cause of loss of the most important resource (manpower). Therefore, it is necessary to improve the preventive processes of disease outbreak to ensure public health. Epidemicity is defined as the unusual occurrence of a disease, event, or behavior which occurs more than predicted (1). Diagnosing and taking action on this issue requires an appropriate context to provide special requirements including appropriate and quick action. Information system is one of the infrastructures with the ability to store, manage process, analyze, and exchange data which is helpful in decision-making. Data storage and data management are two fundamental elements in these systems. One of the important challenges of management in the field of health is the vast amount of unstructured and heterogeneous data which result in the creation of network structures (2). The modeling, storing, managing, processing, and analyzing of such data (especially data related to epidemicity) creates a valuable context for the detection of patterns and other characteristics of contagious diseases. Developing an appropriate data model is the first step in data modeling. A data model is used for the representation of data. In the case of the traditional relational model, data is represented in the form of a table.
However, the most appropriate model for the study of contagious diseases is the graph model. The graph data model consists of nodes and edges. A node is generally used to indicate an entity such as a person in a social network, a place in a transportation system, or a webpage in the internet. The relationship between nodes or entities is shown by edges. Nodes are very similar to the nature of the objects, so object-oriented coding would be close to the data model. Nodes and edges can be made more practical by adding some features on the edges. For example, the node of a disease can contain attributes such as name of disease, description of disease, and the like. Similarly, edges use features to describe connections. For example, the relationship between two individuals in a network can indicate infection and the relation (3). Graph databases can be simply used to answer queries about relationships. Analyzing this network provides important parameters of the disease outbreak such as incidence period, incidence rate, tipping point, prevalence rate, and expected average growth (4). Due to the large volume of data and the complicated structure of the network epidemic model, it is difficult to store data and answer network queries in the database (5, 6).
The term NoSQL is a general name that refers to a set of data models which does not use the structured query language or the relational data model, and it sometimes stands for “Not Only SQL”. This type of system is suitable for working with large amounts of data without the need for relational structure. The term NoSQL was first introduced by Eric Evans in early 2009. Some users of the NoSQL database include Google, Amazon, Twitter, Facebook, and Netflix (7).
The relational data model is one of the models used for medical data in which two kinds of entities exist. One is the main entity which stores the main features of individuals and the other is the dependent entity which stores the features of the disease. This data model has some advantages and disadvantages. One of the advantages is its high efficiency in working with uncomplicated data including short strands, integers, and etc. However, this model does not have the ability to efficiently handle complicated and unstructured data. It is not possible to add attributes while working with this data model. Moreover, this data model cannot support involute data, so it requires many regression procedures to find the disease infection path. This process is time-consuming and can be a waste of time.
Therefore, the data model used in the present study was NoSQL which is a graph database. NoSQL is used to store information because it is able to preserve individuals’ information in nodes and their relationship in edges, and to track information from the graph data model. Since sexual intercourse and infection through some tools have a crucial role in the outbreak of diseases (such as HIV), individuals who transfer the infection should be used in the construction of the data model to better show the infection path. Through the appropriate settings of information in the database, the general data on individuals will be illustrated as a graph. This information can help the analysis of individuals’ relationships and prevention of the infection of healthy individuals.
Graph databases such as Neo4j, an open source database, are used to store data of epidemicity. Edges in the Neo4j model are not only used to show the connection of nodes, but they can also contain some information. This information is stored as value key pairs. This system increases the speed of answering queries by using direct indicators between nodes and edges, and between indexing and relationship. Individuals along with their attributes and laboratory tests are allocated to nodes and edges indicating social relationship among them.
Acquired Immune Deficiency Syndrome (AIDS) was first recognized as a clinical syndrome where healthy individuals were affected by a malignant infection caused by opportunist pathogens. Studies confirm severe immune deficiency with a cellular intermediate in these individuals, and hence, this disease is called Acquired Immune Deficiency Syndrome. When the immune system breaks down, it becomes vulnerable not only to the HIV virus (the first agent causing damage), but also to other infections. The immune system of these individuals is not able to kill any of the microorganisms which previously did not cause any problems. Over time, the infected individuals become more and more ill, and years after the infection, they will develop severe infections or cancers. At this time, it is said that they are infected with HIV. In other words, when a person who is infected with this virus develops a serious disease for the first time, or when the number of immune cells of the body remains below a certain level, this creature is known to be HIV-positive.
Infection with HIV is a serious stage in which the body has a very low defensive power against other infections. In fact, anyone infected with HIV is not necessarily HIV-positive, but he/she can infect others. This process is not visible and there is no way to determine if an individual is HIV-positive by looking at them; it can only be detected by a blood test a few months after the first contact with the virus. Infected individuals may remain in full health for many years and not know about their infection (8). The HIV virus can be transmitted from mother to fetus during intrauterine life or from mother to infant during breastfeeding, and through unprotected sexual contact, shared injection equipment, blood and blood products, and organ/graft transplant.
Individuals along with their relationships, such as unprotected sexual contact (9), shared injection equipment, or transfusion of infected blood, play a crucial role in spreading this disease (10, 11).
Special attention is needed to improve the preventive process which is essential to ensure public health. Various models have been used to store medical data; they were mostly used to record individuals’ medical information at hospitals or health care centers. For this purpose, the relational data model (horizontal tables) is used which has a poor performance in implementing individuals’ relationship and answering the profound level of queries. Moreover, in some cases, this model is not able to track more than three levels. Furthermore, in the relational model, all the attributes should be predetermined and placed in their own column, which is not suitable for our work. For example, due to various attributes, it is not possible to predetermine all the fields, because in some cases, some features are not essential to be noted and this may cause accumulation of basic data in the database. Hence, graph data model, due to its ability to define at the moment, is more practical and optimized. Moreover, according to the graph traversing results, navigation of the profound level would be faster in the graph data model.
The main goal of this paper was to provide an appropriate data model to handle data required for managing HIV-positive patients' data in relative information systems. The most important benefit of the proposed data model is its efficient performance in terms of infection and epidemic detection.
The next section of the paper will provide an overview of previous related works. Then, the proposed data model is presented. The succeeding section of the text presents the performance of the model and discussion about the results. Finally, the paper is concluded and some future works are introduced.
Related work: Thus far, many data models have been used to store medical data, but none of them are the ideal model due to their disadvantages.
One of the issues that communities and organizations are facing is the incidence of new diseases, methods, and changes which significantly affect the health of the society. These changes and the way they are dealt with strongly impact the success and failure of the society. Since the main factor in health care is prevention and quick action after the outbreak of disease, automating these actions would play a significant role in improving the performance of health care providers. Technological changes and disease outbreak are two important issues for governments, the lack of adaptation to which would be harmful to the community, and the cause of loss of the most important resource (manpower). Therefore, it is necessary to improve the preventive processes of disease outbreak to ensure public health. Epidemicity is defined as the unusual occurrence of a disease, event, or behavior which occurs more than predicted (1). Diagnosing and taking action on this issue requires an appropriate context to provide special requirements including appropriate and quick action. Information system is one of the infrastructures with the ability to store, manage process, analyze, and exchange data which is helpful in decision-making. Data storage and data management are two fundamental elements in these systems. One of the important challenges of management in the field of health is the vast amount of unstructured and heterogeneous data which result in the creation of network structures (2). The modeling, storing, managing, processing, and analyzing of such data (especially data related to epidemicity) creates a valuable context for the detection of patterns and other characteristics of contagious diseases. Developing an appropriate data model is the first step in data modeling. A data model is used for the representation of data. In the case of the traditional relational model, data is represented in the form of a table.
However, the most appropriate model for the study of contagious diseases is the graph model. The graph data model consists of nodes and edges. A node is generally used to indicate an entity such as a person in a social network, a place in a transportation system, or a webpage in the internet. The relationship between nodes or entities is shown by edges. Nodes are very similar to the nature of the objects, so object-oriented coding would be close to the data model. Nodes and edges can be made more practical by adding some features on the edges. For example, the node of a disease can contain attributes such as name of disease, description of disease, and the like. Similarly, edges use features to describe connections. For example, the relationship between two individuals in a network can indicate infection and the relation (3). Graph databases can be simply used to answer queries about relationships. Analyzing this network provides important parameters of the disease outbreak such as incidence period, incidence rate, tipping point, prevalence rate, and expected average growth (4). Due to the large volume of data and the complicated structure of the network epidemic model, it is difficult to store data and answer network queries in the database (5, 6).
The term NoSQL is a general name that refers to a set of data models which does not use the structured query language or the relational data model, and it sometimes stands for “Not Only SQL”. This type of system is suitable for working with large amounts of data without the need for relational structure. The term NoSQL was first introduced by Eric Evans in early 2009. Some users of the NoSQL database include Google, Amazon, Twitter, Facebook, and Netflix (7).
The relational data model is one of the models used for medical data in which two kinds of entities exist. One is the main entity which stores the main features of individuals and the other is the dependent entity which stores the features of the disease. This data model has some advantages and disadvantages. One of the advantages is its high efficiency in working with uncomplicated data including short strands, integers, and etc. However, this model does not have the ability to efficiently handle complicated and unstructured data. It is not possible to add attributes while working with this data model. Moreover, this data model cannot support involute data, so it requires many regression procedures to find the disease infection path. This process is time-consuming and can be a waste of time.
Therefore, the data model used in the present study was NoSQL which is a graph database. NoSQL is used to store information because it is able to preserve individuals’ information in nodes and their relationship in edges, and to track information from the graph data model. Since sexual intercourse and infection through some tools have a crucial role in the outbreak of diseases (such as HIV), individuals who transfer the infection should be used in the construction of the data model to better show the infection path. Through the appropriate settings of information in the database, the general data on individuals will be illustrated as a graph. This information can help the analysis of individuals’ relationships and prevention of the infection of healthy individuals.
Graph databases such as Neo4j, an open source database, are used to store data of epidemicity. Edges in the Neo4j model are not only used to show the connection of nodes, but they can also contain some information. This information is stored as value key pairs. This system increases the speed of answering queries by using direct indicators between nodes and edges, and between indexing and relationship. Individuals along with their attributes and laboratory tests are allocated to nodes and edges indicating social relationship among them.
Acquired Immune Deficiency Syndrome (AIDS) was first recognized as a clinical syndrome where healthy individuals were affected by a malignant infection caused by opportunist pathogens. Studies confirm severe immune deficiency with a cellular intermediate in these individuals, and hence, this disease is called Acquired Immune Deficiency Syndrome. When the immune system breaks down, it becomes vulnerable not only to the HIV virus (the first agent causing damage), but also to other infections. The immune system of these individuals is not able to kill any of the microorganisms which previously did not cause any problems. Over time, the infected individuals become more and more ill, and years after the infection, they will develop severe infections or cancers. At this time, it is said that they are infected with HIV. In other words, when a person who is infected with this virus develops a serious disease for the first time, or when the number of immune cells of the body remains below a certain level, this creature is known to be HIV-positive.
Infection with HIV is a serious stage in which the body has a very low defensive power against other infections. In fact, anyone infected with HIV is not necessarily HIV-positive, but he/she can infect others. This process is not visible and there is no way to determine if an individual is HIV-positive by looking at them; it can only be detected by a blood test a few months after the first contact with the virus. Infected individuals may remain in full health for many years and not know about their infection (8). The HIV virus can be transmitted from mother to fetus during intrauterine life or from mother to infant during breastfeeding, and through unprotected sexual contact, shared injection equipment, blood and blood products, and organ/graft transplant.
Individuals along with their relationships, such as unprotected sexual contact (9), shared injection equipment, or transfusion of infected blood, play a crucial role in spreading this disease (10, 11).
Special attention is needed to improve the preventive process which is essential to ensure public health. Various models have been used to store medical data; they were mostly used to record individuals’ medical information at hospitals or health care centers. For this purpose, the relational data model (horizontal tables) is used which has a poor performance in implementing individuals’ relationship and answering the profound level of queries. Moreover, in some cases, this model is not able to track more than three levels. Furthermore, in the relational model, all the attributes should be predetermined and placed in their own column, which is not suitable for our work. For example, due to various attributes, it is not possible to predetermine all the fields, because in some cases, some features are not essential to be noted and this may cause accumulation of basic data in the database. Hence, graph data model, due to its ability to define at the moment, is more practical and optimized. Moreover, according to the graph traversing results, navigation of the profound level would be faster in the graph data model.
The main goal of this paper was to provide an appropriate data model to handle data required for managing HIV-positive patients' data in relative information systems. The most important benefit of the proposed data model is its efficient performance in terms of infection and epidemic detection.
The next section of the paper will provide an overview of previous related works. Then, the proposed data model is presented. The succeeding section of the text presents the performance of the model and discussion about the results. Finally, the paper is concluded and some future works are introduced.
Related work: Thus far, many data models have been used to store medical data, but none of them are the ideal model due to their disadvantages.
- Relational data model for contagious diseases
As is shown in table 1, one of the available data models for storing medical data is the relational data model. There are two entities in this data model. The main entity includes the main features of hospitalized patients such as age, sex, place of birth, and the like. The other is a dependent entity in which the features of the disease are recorded, such as the kind of disease, date of infection, people at risk of infection, the possible transmitter of disease, clinical examination, and laboratory results such as radiography images and etc. Therefore, a database must be able to store different types of data such as images. This data model, however, does not support some types of data including audios and images.
In the process of recording patients’ information, different signs and symptoms may arise. For example, a patient may need to perform a blood sugar test, while another patient may need a liver function test, or in some cases we may encounter symptoms we have never seen before. Accordingly, if all attributes are recorded initially, this model is not able to support the function of insetting additional attributes at any time. Individuals’ relationships are very important in medical data. This model cannot support involute data to find disease infection path, and many regression procedures are needed to find it. This process is time-consuming and would be a waste of time.
In the process of recording patients’ information, different signs and symptoms may arise. For example, a patient may need to perform a blood sugar test, while another patient may need a liver function test, or in some cases we may encounter symptoms we have never seen before. Accordingly, if all attributes are recorded initially, this model is not able to support the function of insetting additional attributes at any time. Individuals’ relationships are very important in medical data. This model cannot support involute data to find disease infection path, and many regression procedures are needed to find it. This process is time-consuming and would be a waste of time.
Table 1: Relational database
| EmployeeID | FirstName | LastName | Age | Salary |
| SM1 | Anuj | Sharma | 45 | 10000000 |
| MM2 | Anand | 34 | 5000000 | |
| T3 | Vikas | Gupta | 39 | 7500000 |
| E4 | Dinesh | Verma | 32 | 2000000 |
- Object-relational data model
One of the recording methods for medical data is Entity Attribute Value with Classes and Relationships (EAV/CR) method. In this method, the object-relational data model is used to store information. This model allows invariable data to be saved in traditional relational databases, and when new data is needed, it uses classes in such a way that each class has its own relational table and columns. The object-relational model can resolve the problem of predetermined scheme and definition of various data formats, but the problem of individuals’ relationships still persists (12).
Figure 1 shows a simple example of EAV/CR method. In this database, many regression queries are likewise needed to find the disease infection path which makes the process time consuming (12).
Figure 1 shows a simple example of EAV/CR method. In this database, many regression queries are likewise needed to find the disease infection path which makes the process time consuming (12).
.jpg)
Figure 1: An overview of the Entity Attribute Value (EAV) (12)
Material and Methods
Individuals create the community together and since HIV virus is transmitted through the body fluid of an infected person to another with the acceptance criteria for the virus, it is very important to detect among whom the disease is transmitted. In other words, gathering the infected individuals in a community provides a general estimation of the transmission of the virus.
Since the population in this study included all people, organizing the genealogy of people and specifying their relationship was important. Moreover, since HIV virus can be transmitted among people with familial relationship, this genealogy will help prevent the outbreak of the disease. For example, a pregnant woman who is a HIV carrier can transfer the virus to her infant during the delivery process; hence, if the disease infection path is known, the pregnancy is prevented or delivery can be performed in a way that virus transmission does not occur.
The population in this data model included individuals’ relationship, the virus infection path, and other related factors. Among the advantages of this data model, finding the ways of infection and introducing a method to show how a healthy individual is affected by a contagious disease such as HIV can be mentioned.
Generally, in this data model, the examination of disease transmission was attempted in the form of the relationship of one person with another as well as by methods other than individuals’ relationships. For example, person A is a healthy person in relationship with person B based on wife-husband or filiation ties. In order to prevent the infection of person A, there would be three modes for person B. In mode 1, person B is completely healthy so the disease is not transmitted through the relationship. In mode 2, person B is infected, so the disease is likely to be transmitted. In mode 3, person B is treated; this mode is unlikely to occur as AIDS has no certain cure and only some preventive methods are available for it.
Elements of the proposed data model: The proposed data model is presented in figure 2. One of the advantages of this data model is that each of the nodes (entities) contains attributes and the edges also show specific features.
Individuals create the community together and since HIV virus is transmitted through the body fluid of an infected person to another with the acceptance criteria for the virus, it is very important to detect among whom the disease is transmitted. In other words, gathering the infected individuals in a community provides a general estimation of the transmission of the virus.
Since the population in this study included all people, organizing the genealogy of people and specifying their relationship was important. Moreover, since HIV virus can be transmitted among people with familial relationship, this genealogy will help prevent the outbreak of the disease. For example, a pregnant woman who is a HIV carrier can transfer the virus to her infant during the delivery process; hence, if the disease infection path is known, the pregnancy is prevented or delivery can be performed in a way that virus transmission does not occur.
The population in this data model included individuals’ relationship, the virus infection path, and other related factors. Among the advantages of this data model, finding the ways of infection and introducing a method to show how a healthy individual is affected by a contagious disease such as HIV can be mentioned.
Generally, in this data model, the examination of disease transmission was attempted in the form of the relationship of one person with another as well as by methods other than individuals’ relationships. For example, person A is a healthy person in relationship with person B based on wife-husband or filiation ties. In order to prevent the infection of person A, there would be three modes for person B. In mode 1, person B is completely healthy so the disease is not transmitted through the relationship. In mode 2, person B is infected, so the disease is likely to be transmitted. In mode 3, person B is treated; this mode is unlikely to occur as AIDS has no certain cure and only some preventive methods are available for it.
Elements of the proposed data model: The proposed data model is presented in figure 2. One of the advantages of this data model is that each of the nodes (entities) contains attributes and the edges also show specific features.
- Entities
After detecting the disease process and evaluating medical documents of HIV-positive patients, the entities (nodes), the relationship, and the attributes in relation to this data model are determined as below:
- Human: National ID card number, full name, date of birth, place of birth, booklet ID number, sex, marital status, place of residence, occupation, email address, and phone number
- Diagnosis: Diagnosis code, and diagnosis explanation
- Patient: Blood pressure, blood group, pulse rate, electrocardiogram, history of the disease, and duration of disease
- Medication: Medication code, medication name, side effects, course of medication, and dosage
- Treatment: Treatment code, treatment explanation and orders, and equipment
- Physician: National ID card number, full name, date of birth, place of birth, booklet ID number, sex, marital status, place of residence, email address, phone number, registration number, registration date, and the date of graduation
- The infection path 1 (Body fluid): Type, explanation, tools, preventive methods, and risk percentage
- The infection path 2 (Shared injection equipment): Type, explanation, tools, preventive methods, and risk percentage
- The infection path 3 (Needlestick): Type of instrument, explanation, tools, preventive methods, and risk percentage
- The infection path 4 (Transfusion of infected blood): Type of blood product, explanation, preventive methods, and risk percentage
.jpg)
Figure 2: The relationship between two nodes and human attributes on the relational edge
- Relationships
The introduced data model is similar to the composition of graph and composition of nodes (entities) and edges (entities’ relationships). Each of the entities mentioned above has relationships with the other corresponding entities which are represented in this section. Directional edges indicate the sequence of entities and show how the information is arranged in the model. Relational edges among nodes and the attributes placed on the edges are classified into the following groups:
- Human-Human edge: This edge shows the relationship between two individuals. Since the graph database has the ability to add attributes to the edges, this edge can also show two common attributes such as the possibility of infection (which is answered with Yes/No), and the kind of relationship between these two individuals. For example, as can be seen in figure 3, on the directional edge labeled Human-Human, the relationship between the two individuals is Mother-Son; this means that human A is the mother and human B is her son and the infection status is yes. The direction of the edge indicates that the disease was transmitted from human A to human B. This system is designed in such a way that, for example, the user enters the primary information about individuals, providing nodes consisting of individuals in the population, and eventually we are directly provided with individuals’ data. Therefore, as soon as we enter information about an individual’s relationship and positive disease transmission, the system provides other information including other entities and connections.
- Diagnosis-Patient edge: Diagnosis is one of the important factors in the cognition of disease. In this model, the diagnosis entity determines whether a healthy person is infected. The edge between these two nodes contains diagnostic factors including HIV-test, blood test, urine analysis and urine culture, chest X-ray, CD4 count, ELISA test, and Western test.
- Physician-Diagnosis edge: This kind of information is recorded by the physician at the time of primary diagnostic measures.
- Patient-Physician edge: This is an ordinary relationship in which some factors are recorded, such as patient’s case number, data filing date, and the number of checkups carried out.
- Physician-Treatment edge: In this relationship, if the infection of the patient is approved, the type of treatment and the course of treatment are determined.
- Physician-Medication edge: After determining the diagnosis and treatment, the type of medications, dosage, and course of consumption are placed between the physician’s and patient’s nodes.
- Patient-Body fluid edge: If infection is diagnosed for a healthy person, then he/she is the entity of a patient. In this case, the infection is transmitted through the patient’s contact with body fluid which indicates a relationship between the patient and one of the infection sources. In this edge, the time of infection, protection status, and duration of exposure to the source of infection are considered.
- Patient-Shared injection equipment edge: The most common route of HIV spreading is using shared injection equipment by injection drug users. Since in this way, the transmission occurs by means of an instrument (syringe), it is important to know the time and place of the incidence to provide necessary preventive measures. Moreover, the condition of the person as a valuable information field is considered.
- Patient-Needlestick edge: For hospital staff including nurses and physicians, the most common route of transmission is blood contact with medical equipment such as a needlestick. Attributes such as the time of occurrence, protection status, and also the place of incidence are considered in this edge.
- Patient-Blood transfusion edge: This edge and the edge of patient-needlestick have the same attributes. Another field called infected blood source is added to this edge so that we can track the spreading of infected blood in health care centers and blood donor centers.
- Patient-Treatment edge: In this edge, the treatment performed earlier or the treatments that should be performed are mentioned, including previous treatment, starting date, effectiveness, and duration.
.jpg)
Figure 3: The proposed graph data model
- Treatment-Body fluid edge: This edge considers the relationship between the type of treatment and the way of transmission by which the patient is infected. Thus, it is clear that the treatment is appropriate to the infection path. A number of factors including starting date, type, and effectiveness of the treatment are considered in this relationship. It should be noted that the edges of treatment-shared injection equipment, treatment-needlestick, and treatment-infected blood transfusion are the same as the treatment-body fluid edge except that the type of treatment is different in each case.
- Treatment-Medication edge: This edge represents the relationship between the treatment and the appropriate medication. This edge indicates that medication is the most important part of treatment and it considers the duration of drug consumption, and its effectiveness duration and side effects.
At the end of this section, it should be noted that the infection paths in this data model are considered as independent entities, so that it would be possible to simply identify the main cause of infection by using information recorded in medical records.
Results
Data preparation: Data sets from the UCI archive (http:/archive.ics.uci.edu) were used to create network epidemic data. This website belongs to the University of Massachusetts and it provides the user with a collection of data of different sciences to put in the tests. Because of the difference in the structure of the incoming data with our data model and our method of data storage, we had to make changes so that they could fit our data model. In addition, as our data model requires the gathering of personal information and their relationship as genealogy, the required information was extracted from the myheritage website (http:/www.myheritage.com). This website receives individuals’ information online and draws their genealogy. After the preparation of data in a genealogic structure and determination of patients’ characteristics, the two required testing and analysis based on the proposed data model.
The prepared information included 200 healthy and sick individuals, and 100 information records to implement any recommended structure including physician, diagnosis, medication, patient, insurance, treatment, and disease infection paths such as body fluid, infected blood infusion, needlestick, and shared injection equipment.
Results
Data preparation: Data sets from the UCI archive (http:/archive.ics.uci.edu) were used to create network epidemic data. This website belongs to the University of Massachusetts and it provides the user with a collection of data of different sciences to put in the tests. Because of the difference in the structure of the incoming data with our data model and our method of data storage, we had to make changes so that they could fit our data model. In addition, as our data model requires the gathering of personal information and their relationship as genealogy, the required information was extracted from the myheritage website (http:/www.myheritage.com). This website receives individuals’ information online and draws their genealogy. After the preparation of data in a genealogic structure and determination of patients’ characteristics, the two required testing and analysis based on the proposed data model.
The prepared information included 200 healthy and sick individuals, and 100 information records to implement any recommended structure including physician, diagnosis, medication, patient, insurance, treatment, and disease infection paths such as body fluid, infected blood infusion, needlestick, and shared injection equipment.
.jpg)
Figure 4: Comparison of graph database and relational database in terms of storage space occupation
System specification: After preparing the data and software, a computer system with hardware and software requirements was selected for the test.
Evaluation results: The performance of the system was evaluated in the conventional data model and proposed data model using practical and comparative methods in terms of memory consumption for data storage, and the required time to perform queries to find the infection
path.
As is shown in figure 4, the graph database has better performance compared to the relational data model in such a way that by entering the same information in both databases, the graph database occupies less space for data storage. As is clear in the diagram, for data volume at a rate of 100 records, the difference between the two databases in terms of space occupation for each entity and edge was 400 megabytes.
Evaluation results: The performance of the system was evaluated in the conventional data model and proposed data model using practical and comparative methods in terms of memory consumption for data storage, and the required time to perform queries to find the infection
path.
As is shown in figure 4, the graph database has better performance compared to the relational data model in such a way that by entering the same information in both databases, the graph database occupies less space for data storage. As is clear in the diagram, for data volume at a rate of 100 records, the difference between the two databases in terms of space occupation for each entity and edge was 400 megabytes.
Table 2: Comparison of the data retrieval time for different queries in the graph data model and the relational data model
| Query | Graph (response time) |
Relational (response time) |
|
| 1 | Human (demographic) information | 126 | 18 |
| 2 | Information about the physician | 190 | 30 |
| 3 | Information about medications and side effects | 64 | 18 |
| 4 | Information about infection paths | 66 | 14 |
| 5 | Information about a specific physician | 272 | 33 |
| 6 | Information about a specific medication or treatment | 260 | 19 |
| 7 | Information about a specific insurance and a specific patient | 240 | 16 |
| 8 | Information about a specific infection path | 252 | 22 |
| 9 | Information about a specific diagnosis | 270 | 17 |
| 10 | Information about a specific treatment | 290 | 21 |
| 11 | Number of infected individuals in the community | 96 | 7 |
| 12 | Individuals who play the key role in the infection | 720321 | 17250 |
| 13 | Number of individuals who are infected by a specific person | 90 | 150 |
| 14 | Prediction of individuals who are most at risk of infection | 39526 | 162991 |
| 15 | Infection paths to the level 4 | 45 | 920 |
| 16 | The shortest distance between two nodes | 652 | - |
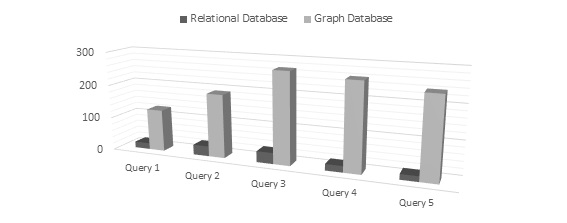
The results of queries in the network relational field which has been executed in the relational and graph models are shown in table 2. Comparison of the diagrams in terms of time shows that the retrieval time of queries for one and two nodes in the graph data model was much longer than the relational database (Figure 5).

Figure 5: Comparison of the run time of queries for one and two nodes in the graph database and relational database
Another issue is finding the source of infection. To achieve this goal, it is necessary to find the infection path. In the proposed graph data model, this would be gained simply by tracking the edges of infection, while in the relational data model, many regression procedures are needed which make it difficult to use. The time consumed to find the infection path to the level of four was 45 milliseconds for the graph model, and 920 milliseconds for the relational model. By increasing the level of infection path, the performance of the graph model increased compared to the relational model.
One of the important queries in this field is finding people who are at risk of the disease. To answer this question, it is necessary to find individuals located in the neighborhood of the infected individuals or at a certain distance from them. In the graph model, this only requires tracking to the desired depth of the graph. However, in the relational model, a connection to a large number of tables is needed and this takes a long time. For example, in the graph data model, it takes 8540 milliseconds to achieve a depth of four, while it takes 2354921 milliseconds in the relational data model (about 276 times longer than the required time in the graph data model). Moreover, the relational model cannot respond to tracking of more than level four.
Another feature of this graph data model is the capability to display all the nodes, edges, and attributes at the same time (row 4 of table 2), but this feature is not available in relational databases.
Discussion
As was mentioned, diseases have been of critical concern to societies and their manpower. Thus, it is essential to accumulate information about diseases and their preventive methods, especially viral diseases. To this purpose, a structure is required to store data in a specific format by which correct analysis is possible. In this study, the graph structure (data model) was used to store data, in which individuals’ features are recorded in the nodes as attributes and edges represent social relationships between them. In the case of transmission of infection from one person to another, the infection attribute becomes "true" for them. After identifying the disease, other entities with their own features and attributes will be added; these entities include infection path, physician, and medication. This model has high performance in answering the queries related to individuals’ relationships such as the disease infection path and people at risk of the infection. Nevertheless, the relational model has a better function only in queries related to data retrieval.
This system can be improved by the combination of the graph model and other models so that it would be able to effectively answer both types of queries (retrieval of individuals’ features and tracking of their relationships). For example, the combination of graph and relational models is an appropriate choice to store network epidemic data. Moreover, the focus of this study was static social networks. Hence, it is important to offer a model to store dynamic social networks such as individuals’ communication patterns, time, and location.
Conclusion
The proposed system can quickly provide the time and onset of disease transmission on different levels of individuals’ relationships. It makes possible to determine the one beginning point of disease transmission in a specific region of society, and the way by which transmission has occurred. These reports and related analyses can effectively help prevent disease incidence and viral transmission route in epidemic diseases. Using the graph data model allows us to make changes in information at running time, even if the necessary data are not considered while designing a process which is not feasible in other systems.
The management of data for epidemic detection of HIV infection requires an appropriate data model that can provide the required functionalities and features with an acceptable quality. Graph data models are suitable NoSQL models for some of these features (e.g., epidemic detection via traversing of the graph). The proposed graph-based data model provides the main functionalities and features while outperforming performance and utilization metrics.
Acknowledgments
We would like to thank our colleagues at Tarbiat Modares University and Islamic Azad University (Qeshm Island Unit).
Conflict of Interest: None declared.
References
1. Hatemi H, Razavi SM, Eftekhar H. Comprehensive volume of Public Health. 2nd ed. Tehran: Arjomand Publication; 2012. Chapter 8, Part 12, Investigation and Control of Epidemics; P.1010-22.
2. Hey T, Tansley S, Tolle K. The Fourth Paradigm: Data-Intensive Scientific Discovery. 1st ed. New York, United States of America: Microsoft Research; 2009. P.91-134.
3. Stattner E, Vidot N. Social network analysis in epidemiology: Current trends and perspectives. Paper presented at: The 5th International Conference on Research Challenges in Information Science (RCIS), 2011 May 19-21; Gosier, Guadeloupe.
4. Salathe M, Jones JH. Dynamics and control of diseases in networks with community structure. PLoS Comput Biol 2010; 6(4):1-11.
5. Han J, Haihong E, Le G, Du J. Survey on NoSQL database. Paper presented at: The 6th International Conference on Pervasive Computing and Applications (ICPCA); 2011 Oct 26-28; Port Elizabeth, South Africa.
6. Leavitt N. Will NoSQL databases live up to their promise? Computer 2010; 43(2):12-4.
7. Strauch Ch. NoSQL databases. Lecture Selected Topics on Software-Technology Ultra-Large Scale Sites, Stuttgart Media University. 2011. 149 p. Available from: www.christof-strauch.de/nosqldbs.pdf
8. Powers KA, Ghani AC, Miller WC, Hoffman IF, Pettifor AE, Kamanga G, et al. The role of acute and early HIV infection in the spread of HIV and implications for transmission prevention strategies in Lilongwe, Malawi: a modelling study. Lancet 2011; 378(9787):256-68.
9. Miller WC, Rosenberg NE, Rutstein SE, Powers KA. The role of acute and early HIV infection in the sexual transmission of HIV. Curr Opin HIV AIDS 2010; 5(4):277-82.
10. Basavaraj KH, Navya MA, Rashmi R. Quality of life in HIV/AIDS. Indian J Sex Transm Dis 2010; 31(2):75-80.
11. Levinson W, Jawetz E. Medical microbiology and immunology: examination & board review. 8th ed. New York, N.Y.: Lange Medical Books/Mc Graw -Hill; 2004.
12. EI-Sappagh ShH, El-Masri S; Riad AM; Elmogy M. Electronic health record data model optimized for knowledge discovery. International Journal of Computer Science Issues 2012; 9(5):329.
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution 4.0 International License. |





